张宇,复旦大学生殖与发育研究院/附属妇产科医院青年研究员,国家级青年人才项目获得者,获国家自然科学基金面上、青年基金、国家重点研发计划、上海市面上、启明星扬帆等项目资助。集中于表观遗传调控配子发生与早期胚胎发育并介导子代慢病易感性机制研究,长期利用分子生物学、发育生物学和计算生物学的方法,探索表观遗传信息在配子发生及早期胚胎发育中的动态变化、作用机制及其生物学功能,并解析其代际及跨代遗传分子机制,共发表高水平SCI论文30余篇,其中包括以第一及通讯作者(含共同)在Nature Genetics、Nature Cell Biology、Molecular Cell、Nature Structural & Molecular Biology、Science Advances、Nature Communications、Cell Discovery等发表论文17篇,研究成果获领域内广泛关注与认可,总引用数超过2300次(谷歌学术)。
主要研究方向 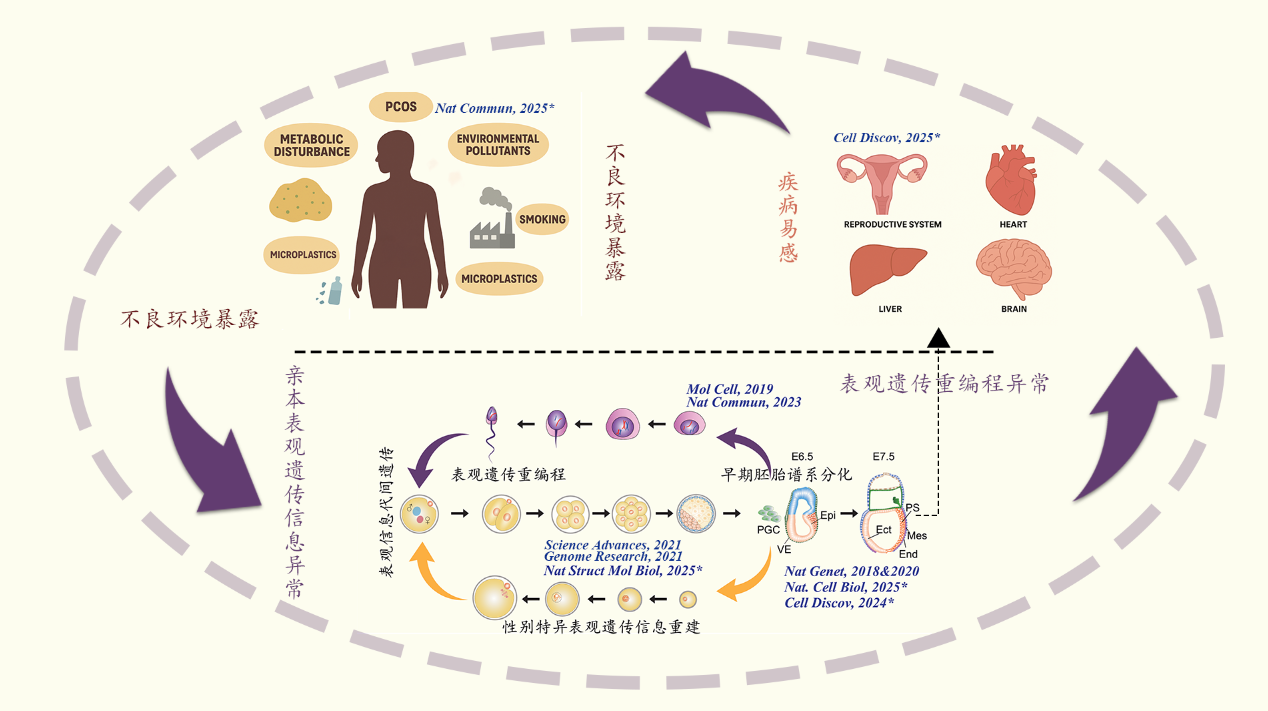
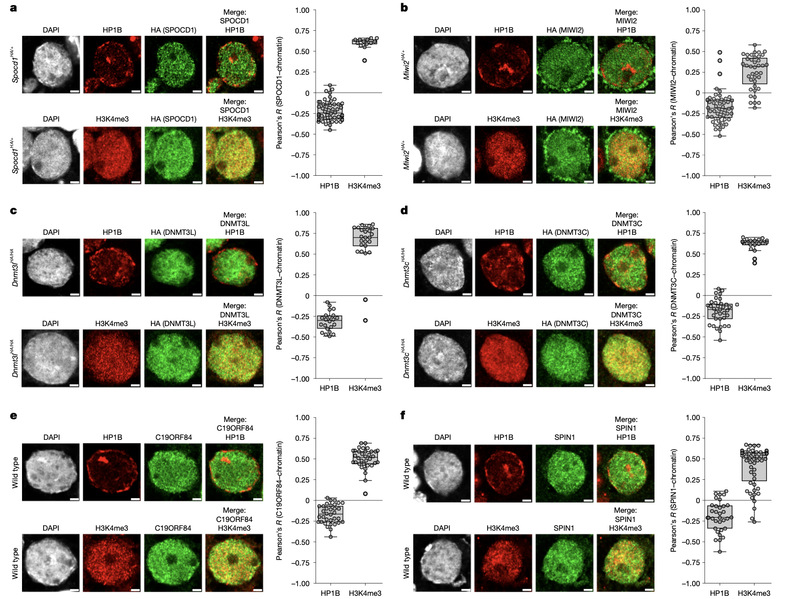
1. 亲本表观遗传信息的建立及其代间传递机制 父本配子表观遗传信息建立及动态变化:解析了恒河猴及小鼠精子发生各阶段染色质高级结构动态变化(Molecular Cell, 2019),并受邀为Current Opinion in Genetics & Development撰写review。随后,我们在不育患者体内发现CCER1基因突变,并利用敲除小鼠发现该基因突变后染色质高级结构改变、组蛋白-鱼精蛋白转换异常并最终导致男性不育(Nature Communications, 2023)。 母本配子表观遗传信息建立及代间传递:构建了人类早期胚胎全基因组亲本特异甲基化图谱(Genome Research, 2021);进一步收集了大鼠、猪、牛的卵子及早期胚胎表观遗传数据集,通过整合分析揭示卵子基因印记建立同时具有物种保守性与物种特异性,尽管不同物种中表观遗传信息的遗传和重编程的模式不同,但是它们都围绕解决印记区的建立和非印记区(如基因转录调控区域)的保护这一“该不该甲基化”(to-methylate-or-not)的问题中实现了有效的表观遗传组的建立和重编程(Science Advances, 2021);发现H2A.Z参与卵子顺式调控元件建立乙酰化(活性)建立,其缺失导致启动子和增强子的活性受损影响关键基因表达,阻碍卵母细胞减数第二次分裂恢复并导致胚胎着床前死亡(Nature Structural & Molecular Biology, 2025)。 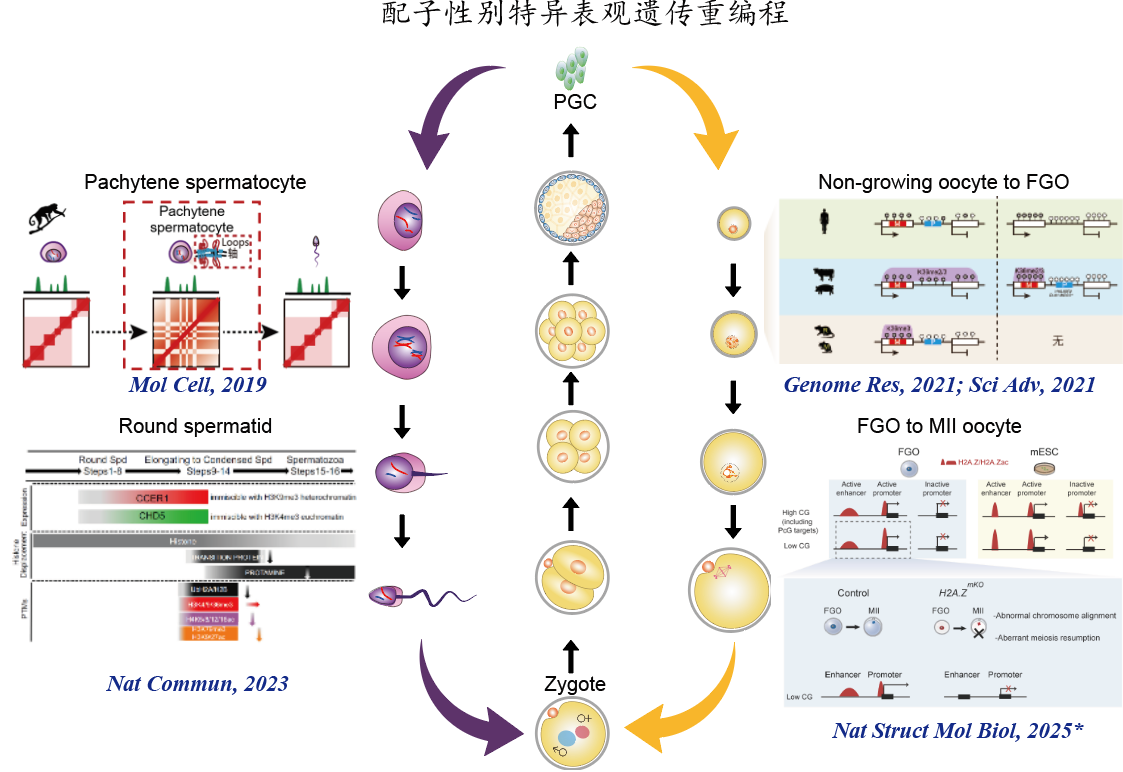
2.表观遗传信息调控早期胚胎谱系分化的机制探索 开发了全新的微量细胞全基因组单碱基精度甲基化建库方法STEM-seq;系统地描绘了哺乳动物中早期胚胎谱系分化及细胞命运决定过程中转录组、DNA甲基化组动态变化,并深入探索了DNA甲基化参与早期谱系细胞分化的调控机制(Nature Genetics, 2018);发现早期胚胎H3K36me2“两步法”建立模式,进一步揭示了H3K36me2协同dosage-dependent DNMT3A/B调控着床后谱系特异差异甲基化模式建立的分子机制(Nature Cell Biology, 2025);利用STAR-ChIP-seq、ATAC-seq及sisHi-C,发现原肠运动‘始发态(primed)’多能性细胞具有特殊染色质状态super-bivalency,且利用基因编辑小鼠证明其对哺乳动物早期胚胎发育至关重要(Nature Genetics, 2020);同时,申请人利用空间转录组技术Stereo-seq构建了E7.5到E14.5完整小鼠胎盘时空转录组图谱,揭示了滋养层发育和胎盘血管生成的关键调节因子并发现印记基因在胎盘呈现空间区域状(compartmentalized)差异分布模式(Cell Discovery, 2024)。 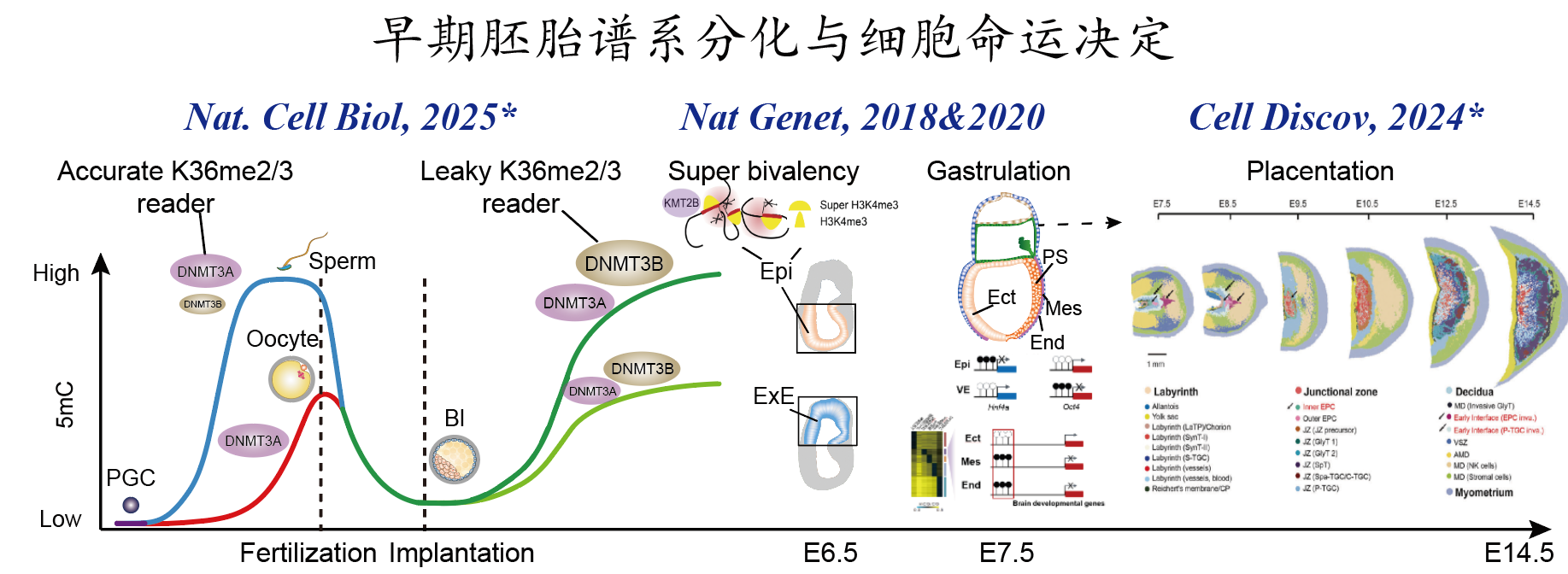
3. 亲代不良环境暴露经表观遗传调控子代发育及健康的分子机制 亲本代谢异常暴露导致子代发育异常:利用小鼠模型揭示宫内高糖(妊娠期糖尿病)通过改变PGC相关代谢产物如α-KG,干扰雌性PGC染色质可及性重编程,导致多能性网络关闭异常、减数分裂程序启动迟滞并最终致其生育力下降的分子机制(Cell Discovery, 2025)。 多囊卵巢综合征发病及代间遗机制探索:利用临床样本及小鼠模型,申请人发现PCOS 女性卵泡发育微环境中脂肪酸利用率增加及表观遗传修饰改变(J Nutr Biochem, 2024);利用临床与小鼠模型证实高雄激素上调颗粒细胞SLC1A5驱动Gln来源的α-KG异常累积,α-KG通过HDAC5介导的H3K14ac/H3K56ac去乙酰化抑制芳香化酶CYP19A1转录加剧高雄激素血症并形成恶性循环参与PCOS(Nature Communications, 2025)。 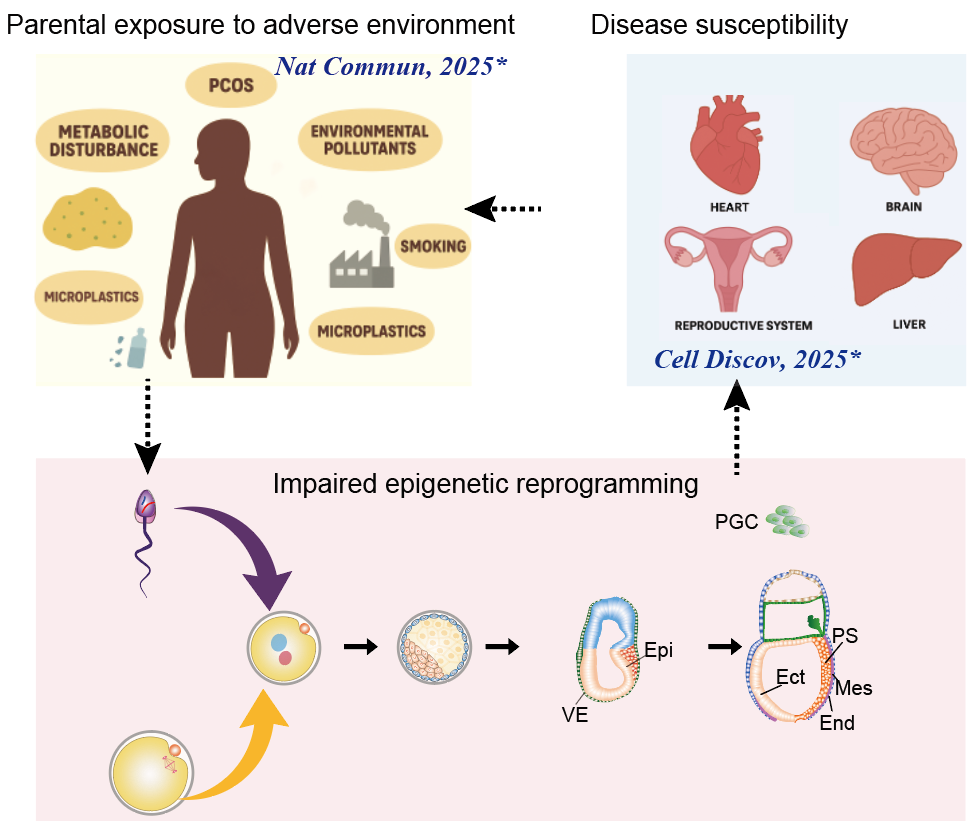
主要科研项目 国家自然科学基金委员会面上项目,国家自然科学基金委员会,2024-2027,主持; 国家自然科学基金委员会青年科学基金项目,国家自然科学基金委员会,2024-2026,主持; 上海市科学技术委员会“科技创新行动计划”自然科学基金面上项目,上海市科学技术委员会,主持; 上海市科学技术委员会 “科技创新行动计划”启明星培育(扬帆专项),上海市科学技术委员会,主持; 国家重点研发计划子课题:葡萄胎发生和恶变的机制及队列研究,科技部,骨干。
发表论文(#第一及共同第一作者,*通讯及共同通讯作者) 2025年 Lu XK#*, Wang LJ#, Liu BF, Hu XY, Wang ZM, Liu L, Yu G, Dong LJ, Kong F, Fan Q, Zhang Y*, Xie W*. Reprogramming of H3K36me2 guides lineage-specific post-implantation de novo DNA methylation. Nature Cell Biology. 2025. Xu QH#, Huang CY#, Ming J, Lu XK, Liu L, Du ZH, Chen Z, Na J, Li GH, Xiang YL*, Zhang Y*, Xie W*. H2A.Z is essential for oocyte maturation and fertility in female mouse. Nature Structural & Molecular Biology. 2025. Cong JS#, Li Q#*, Li YY#, Li MH#, Shi Y, Hu PR, Yin XD, Zhang QY, Sheng JZ, Li JS, Ding GL*, Zhang Y*, Huang HF*. Intrauterine hyperglycemia impairs mouse primordial germ cell development and fertility by sex-specific epigenetic reprogramming interference. Cell Discov. 2025. Wang YS#, Wu JY#, Zhang GC, Shi Y, Meng YC, Lv PP, Huang WW, Su YF, Zhou ZY, Wang B, Chen XJ, Zhou CL, Pan JX, Jin L, Wang XT, Wu YT, Sheng JZ, Liu XM, Zhang Y*, Ding GL*, Yu CJ*, Huang HF*. Androgens drive SLC1A5-dependent metabolic 1 reprogramming in polycystic ovary syndrome. Nat Commun. 2025. Gan S#*, Zhou S#, Zhou J#, Zhang G#, Chen J, Liu R, Sun K, Li S, Xiong W, Wen Y, Sheng J, Zhang Y, Ren J*, Li Y*, Huang H*, Zhang C*. Therapeutic Repair of Sperm Quality Decline Caused by Polytetrafluoroethylene. Adv Sci (Weinh). 2025 Cui J#, Wang C#, Zheng Y, Zhang Y, Luo S, Ren Z, Qin X, Zhang M, Gao F, Huang H, Shu J*, Ding G*. Mechanisms and reversibility of nicotine-induced spermatogenesis impairment and DNA methylation changes. Commun Biol. 2025
2024年 Wu Y#, Su K#, Zhang Y#, Liang L#, Wang F, Chen S, Gao L, Zheng Q, Li C, Su Y, Mao Y, Zhu S, Chai C, Lan Q, Zhai M, Jin X, Zhang J, Xu X, Zhang Y*, Gao Y*, Huang H*. A spatiotemporal transcriptomic atlas of mouse placentation. Cell Discov. 2024. Xu Y#, Zhou Z#, Zhang G, Yang Z, Shi Y, Jiang Z, Liu Y, Chen H, Huang H*, Zhang Y*, Pan J*. Metabolome implies increased fatty acid utilization and histone methylation in the follicles from hyperandrogenic PCOS women. J Nutr Biochem. 2024. Luo QY#, Zhang SW#, Wu HY, Mo JY, Yu JE, He RK, Jiang ZY, Zhu KJ, Liu XY, Lin ZL, Sheng JZ, Zhang Y*, Wu YT*, Huang HF*. Safety of embryo cryopreservation: insights from mid-term placental transcriptional changes. Reprod Biol Endocrinol. 2024. Huang X#*, Balmer S, Lyu C, Xiang Y, Malik V, Wang H, Zhang Y, Cai B, Xie W, Hadjantonakis AK, Zhou H, Wang J*. ZFP281 controls transcriptional and epigenetic changes promoting mouse pluripotent state transitions via DNMT3 and TET1. Dev Cell. 2024
2023年 Qin D#, Gu Y#, Zhang Y#, Wang S#, Jiang T, Wang Y, Wang C, Chen C, Zhang T, Xu W, Wang H, Zhang K, Hu L, Li L, Xie W*, Wu X*, Hu Z*. Phase-separated CCER1 coordinates the histone-to-protamine transition and male fertility. Nat Commun. 2023. Zhang G, Mao Y, Zhang Y, Huang H*, Pan J*. Assisted reproductive technology and imprinting errors: analyzing underlying mechanisms from epigenetic regulation. Hum Fertil (Camb). 2023
2022年及以前(仅展示一作及通讯文章) Zhang Y#, Xiang YL#, Yin QZ#, Du ZH#, Peng X, Wang QJ, Fidalgo M, Xia WK, Li YY, Zhao ZA, Zhang WH, Ma J, Xu F, Wang JL, Li L, Xie W. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nature Genetics. 2018. Wang Y#, Wang H#, Zhang Y#, Du ZH#, Si W#, Fan SX, Qin DD, Wang M, Duan YC, Li LF, Jiao YY, Li YY, Wang QJ, Shi QH, Wu X*, Xie W*. Reprogramming of Meiotic Chromatin Architecture during Spermatogenesis. Molecular Cell. 2019. Xiang YL#, Zhang Y#, Xu QH, Zhou C, Liu B, Du ZH, Zhang K, Zhang BJ, Wang XX, Gayen S, Liu L, Wang Y, Li YY, Wang QJ, Kalantry S, Li L, Xie W. Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency. Nature Genetics. 2020. Lu XK#, Zhang Y#, Wang LJ#, Wang LY#, Wang HL, Xu QH, Xiang YL, Chen CL, Kong F, Xia WK, Lin ZL, Ma SN, Liu L, Wang XG, Ni HM, Li W*, Guo Y*, Xie W*. Evolutionary epigenomic analyses in mammalian early embryos reveal species-specific innovations and conserved principles of imprinting. Science Advances. 2021. Xu JW#, Shu YM#, Yao GD#, Zhang Y#, Niu WB, Zhang YL, Ma XS, Jin HX, Zhang FL, Shi SL, Wang Y, Song WY, Dai SJ, Cheng LY, Zhang XY, Xie W*, Hsueh Aaron J*, Sun YP*. Parental methylome reprogramming in human uniparental blastocysts reveals germline memory transition. Genome Research.2021. Liu YH#, Xu Z#, Shi JJ#, Zhang Y#, Yang ST#, Chen Q, Song CL, Geng SH, Li Q, Li JS, Xu GL, Xie W, Lin HD, Li XJ. DNA methyltransferases are complementary in maintaining DNA methylation in embryonic stem cells. iScience. 2022. Zhang Y and Xie W*. Building the Genome Architecture during Maternal to Zygotic Transition. Current Opinion in Genetics & Development. 2022. Xu Zhen#, Shi Jiajia#, Zhang Yu#, Liu Yuhan, Zhao Junzheng, Chen Qian, Song Chenglin, Geng Shuhui, Xie Wei, Wu Feizhen, Bai Yun, Yang Yang, Li Xiajun. Zfp57 exerts maternal and sexually dimorphic effects on genomic imprinting. Frontiers in Cell and Developmental Biology, 2022. Mo JY, Liu XQ, Huang YT, He RK, Zhang Y*, Huang HF*. Developmental origins of adult diseases. Medical review. 2022.
代表性著作 1.《承续的魅力:令人着迷的表观遗传学》 表观遗传信息的遗传与重编程,科学出版社,2018,参与编写.
获奖 1.第五届海聚英才全球创新创业大赛(省市级)优胜奖,参与,第三完成人,2025.11;
学术组织兼职 1.中国遗传学会表观遗传分会,委员; 2.中国生物信息学学会表观遗传信息分委会,委员 |

 EN
EN